UCLA Fielding School of Public Health researcher’s work opens doors to understanding COVID-19’s early spread
Dr. Marc Suchard co-led two related studies that found early interventions were effective at stamping out infections before they spread.
A UCLA Fielding School of Public Health researcher’s work on two related research projects published in the past month suggests that in both the United States and in Europe, sustained transmission networks of SARS-CoV-2 became established only after separate introductions of the virus that went undetected.
“Our research shows that when you do early intervention and detection well, it can have a massive impact, both on preventing pandemics and controlling them once they progress,” said Dr. Marc Suchard, professor of biostatistics, biomathematics, and human genetics. “While the epidemic eventually slipped through, there were early victories that show us the way forward: Comprehensive testing and case identification are powerful weapons.”
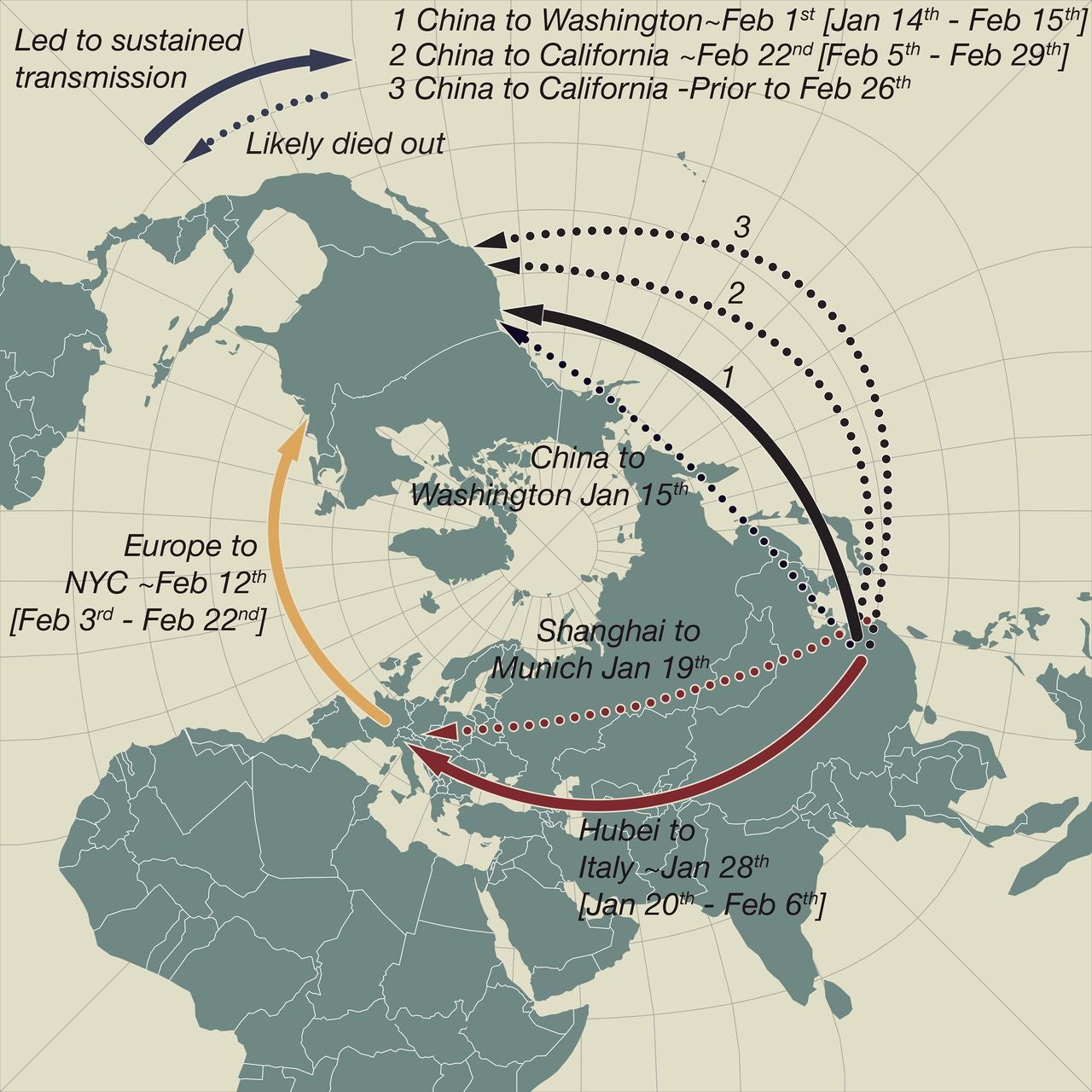 This schematic map shows early and apparently ‘dead-end’ introductions of SARS-CoV-2 to Europe and the US (dashed arrows). Successful dispersals between late January and mid-February are shown with solid arrows: from Hubei Province, China to Northern Italy, from China to Washington State, and later from Europe (as the Italian outbreak spread more widely) to New York City and from China to California. (Illustration by Andrew Rambaut/University of Edinburgh and Jeffrey Joy/University of British Columbia).
This schematic map shows early and apparently ‘dead-end’ introductions of SARS-CoV-2 to Europe and the US (dashed arrows). Successful dispersals between late January and mid-February are shown with solid arrows: from Hubei Province, China to Northern Italy, from China to Washington State, and later from Europe (as the Italian outbreak spread more widely) to New York City and from China to California. (Illustration by Andrew Rambaut/University of Edinburgh and Jeffrey Joy/University of British Columbia).
Suchard’s work as a biostatistician underlies the two related research projects, one published this week in the Nature Research journal Nature Communications, and another published by the American Association for the Advancement of Science (AAAS) journal Science in September.
Specifically, the work combines evolutionary genomics from coronavirus samples with computer-simulated epidemics and detailed travel records to reconstruct the spread of coronavirus across the world, and in unprecedented detail. The teams include scientists from 13 research institutions in the U.S., Belgium, Canada and the United Kingdom.
"Our aspiration was to develop and apply powerful new technology to conduct a definitive analysis of how the pandemic unfolded in space and time, across the globe," said University of Arizona researcher Michael Worobey, who worked on both of the teams with Suchard. “Before, there were lots of possibilities floating around in a mish-mash of science, social media and an unprecedented number of preprint publications still awaiting peer review."
That effort, whose findings were published in Science, based their analysis on results from viral genome sequencing efforts, which began immediately after the virus was identified. These efforts quickly grew into a worldwide effort unprecedented in scale and pace and have yielded tens of thousands of genome sequences, publicly available in databases.
Contrary to widespread narratives, the first documented arrivals of infected individuals traveling from China to the U.S. and Europe did not snowball into continental outbreaks, the researchers found.
“Swift and decisive measures aimed at tracing and containing those initial incursions of the virus were successful,” Suchard said. “These should serve as model responses directing future actions and policies by governments and public health agencies.”
The team also detailed how they reconstructed the pandemic's unfolding by running computer programs that carefully simulated the epidemiology and evolution of the virus – essentially, how SARS-CoV-2 spread and mutated over time.
"This allowed us to re-run the tape of how the epidemic unfolded, over and over again, and then check the scenarios that emerge in the simulations against the patterns we see in reality," Worobey said.
In the second study, published this month by Nature Communications, new methods were then combined with the data from the epidemic reconstruction, yielding exceptionally detailed and quantitative results.
"Fundamental to this work stands our new tool combining detailed travel history information and phylogenetics, which produces a sort of 'family tree' of how the different genomes of virus sampled from infected individuals are related to each other," Suchard said. “The more accurate evolutionary reconstructions from these tools provide a critical step to understand how SARS-CoV-2 spread globally in such a short time.”
Funding:
Funding sources for these studies include the David and Lucile Packard Foundation, the National Institutes of Health, the European Research Council, the Wellcome Trust and the Canadian Institutes of Health Research Coronavirus Rapid Response Programme.
Citations:
Science – Science 10 Sep 2020: eabc8169; DOI: 10.1126/science.abc8169
Nature Communications - Nat Commun 11, 5110 (2020). https://doi.org/10.1038/s41467-020-18877-9

