Biostatistics Seminars
Winter 2021
-
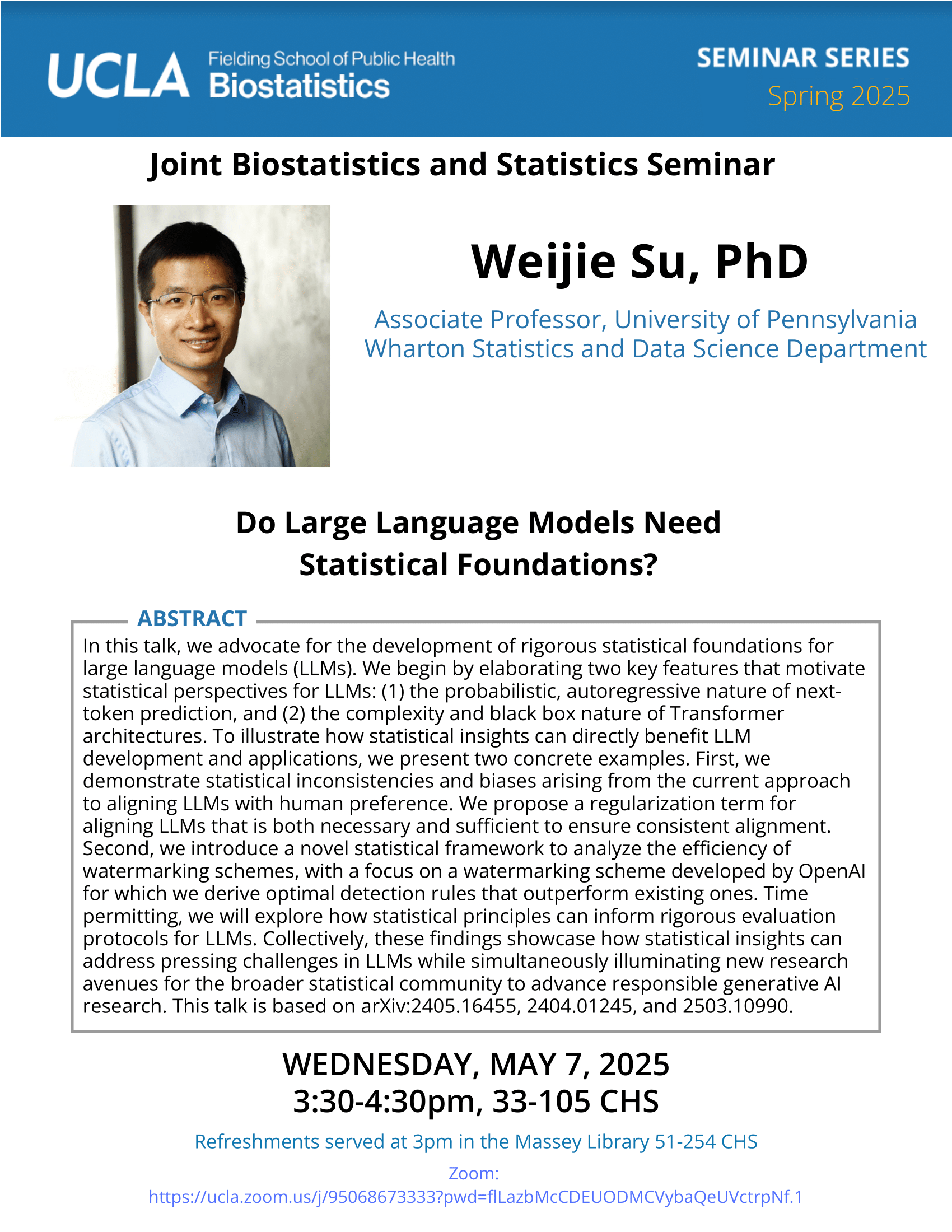
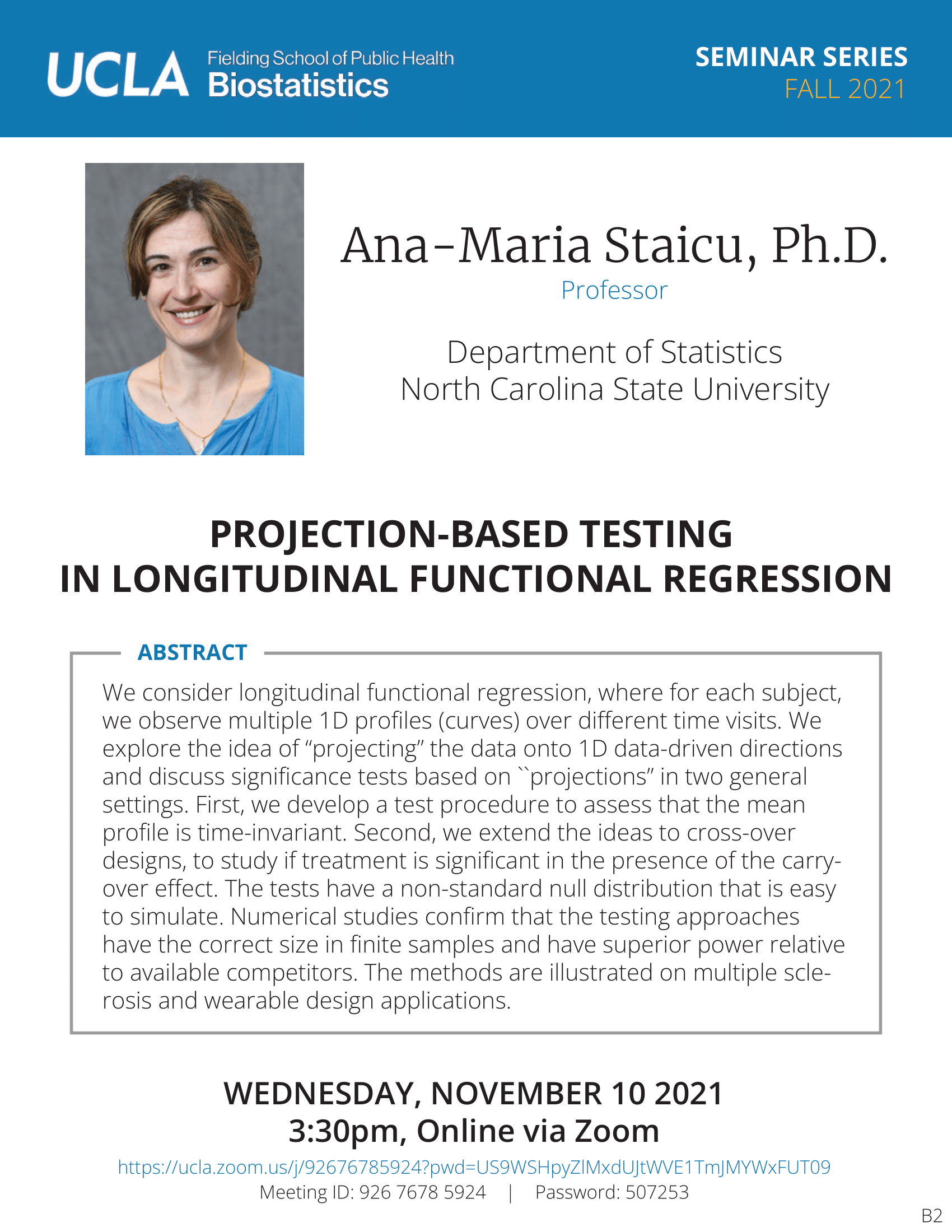
Feifei Xiao - 3/8/21
Identification and Characterization of Genomic Variants with High Throughput Data | UCLA CTSI Biostatistics Seminar
Date
Monday, March 8, 2021Time
12:00pm - 1:00pmLocation
Online via Zoom
https://uclahs.zoom.us/j/780633065Speaker
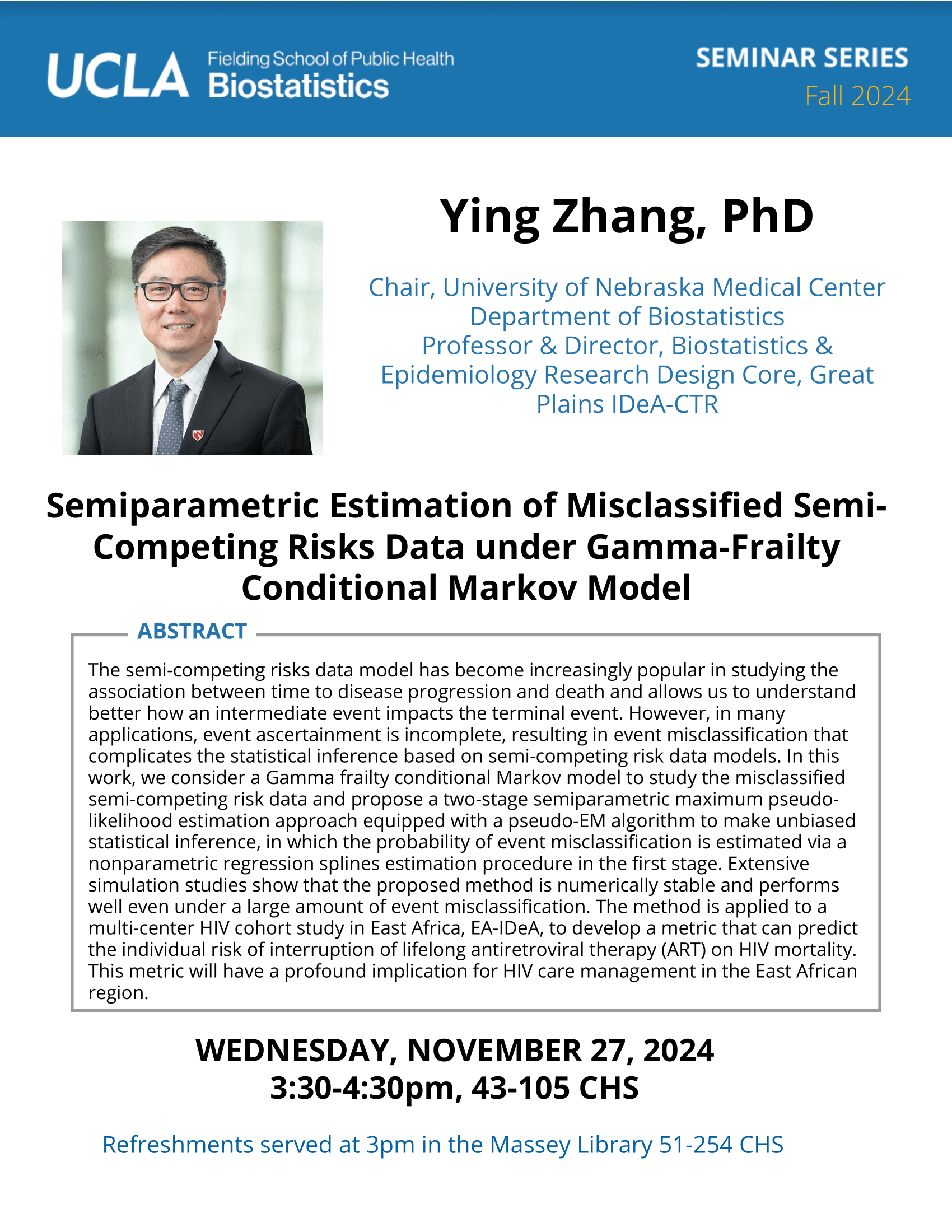
Feifei Xiao, Ph.D.
Associate Professor, University of South CarolinaBio
Feifei Xiao, Ph.D, is an Assistant Professor in the Department of Epidemiology and Biostatistics at the University of South Carolina. Dr. Xiao received her Ph.D. in Biostatistics from The University of Texas MD Anderson Cancer Center in 2013. She then got her postdoc training in Biostatistics from School of Public Health at Yale University (2013-2015). Dr. Xiao’s research focuses on high throughput genetic/genomics data, specifically on copy number variations, gene-gene/environment interactions, epigenetics and next generation sequencing data analysis. She has published 27 articles in peer reviewed journals of statistics, genetics and bioinformatics including Nucleic Acid Research, Human Genetics, and Bioinformatics.Abstract
Massive datasets generated by modern technologies have enabled great effort toward precision medicine. Researchers have identified various genetics/genomics features as potential biomarkers for disease prevention and diagnosis. The first part of my talk will be on copy number variants (CNVs) analysis. Most of existing methods used algorithms assuming that the observed data of different genetic loci are independent. Our study found that the correlation structure of CNV data is associated with linkage disequilibrium. Therefore, we developed a novel algorithm that will systematically integrate the genomic correlation structure into the modeling. I will show simulations and the application to a whole genome melanoma study. Application to a large cohort lung cancer study to reveal high confidence CNVs predisposing to lung cancer risk will also be illustrated. In the second part of my talk, I will talk about the identification of a gene expression based immune signature for lung adenocarcinoma prognosis using machine learning methods.Participating CTSI Institutions
UCLA, Harbor-UCLA, Charles Drew University, and Cedars-Sinai -
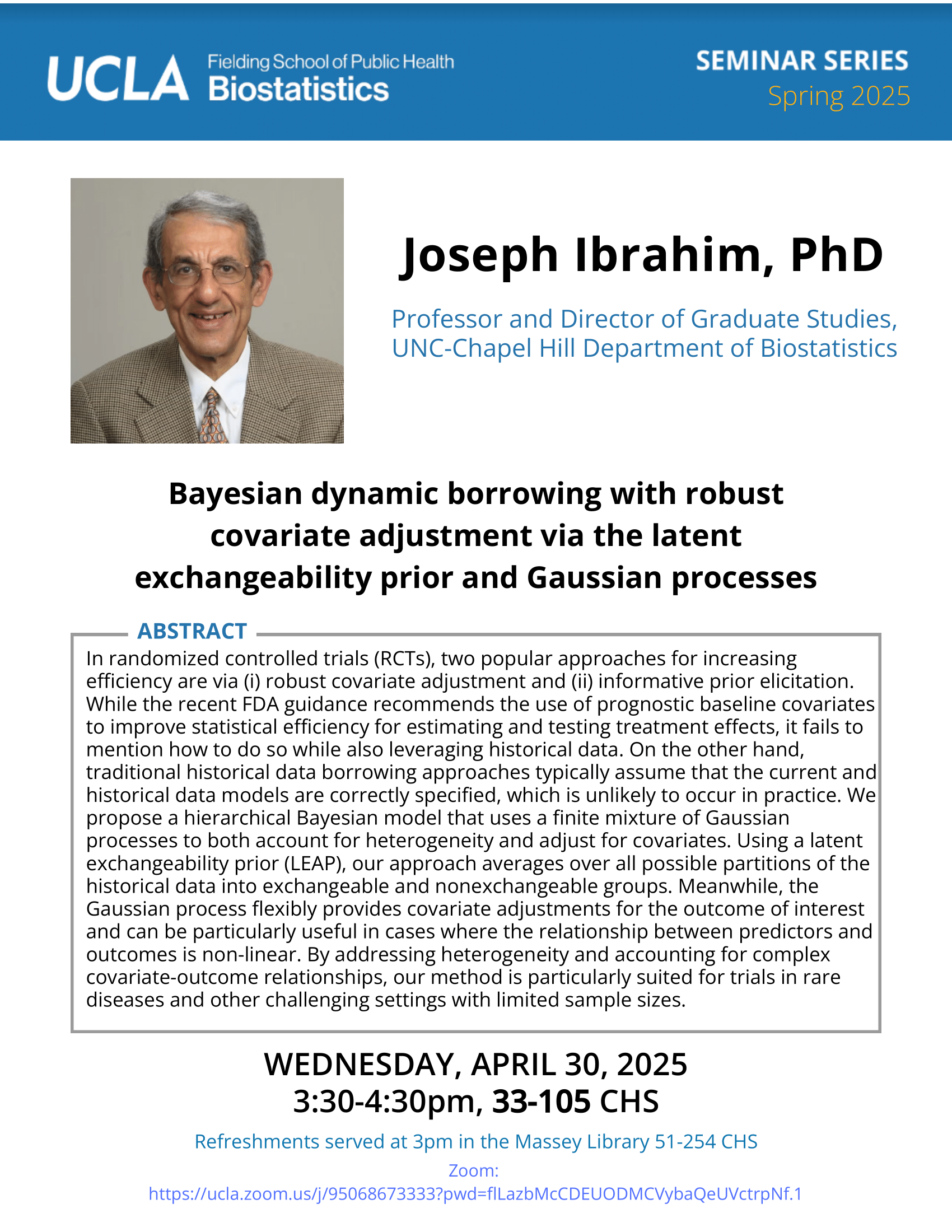
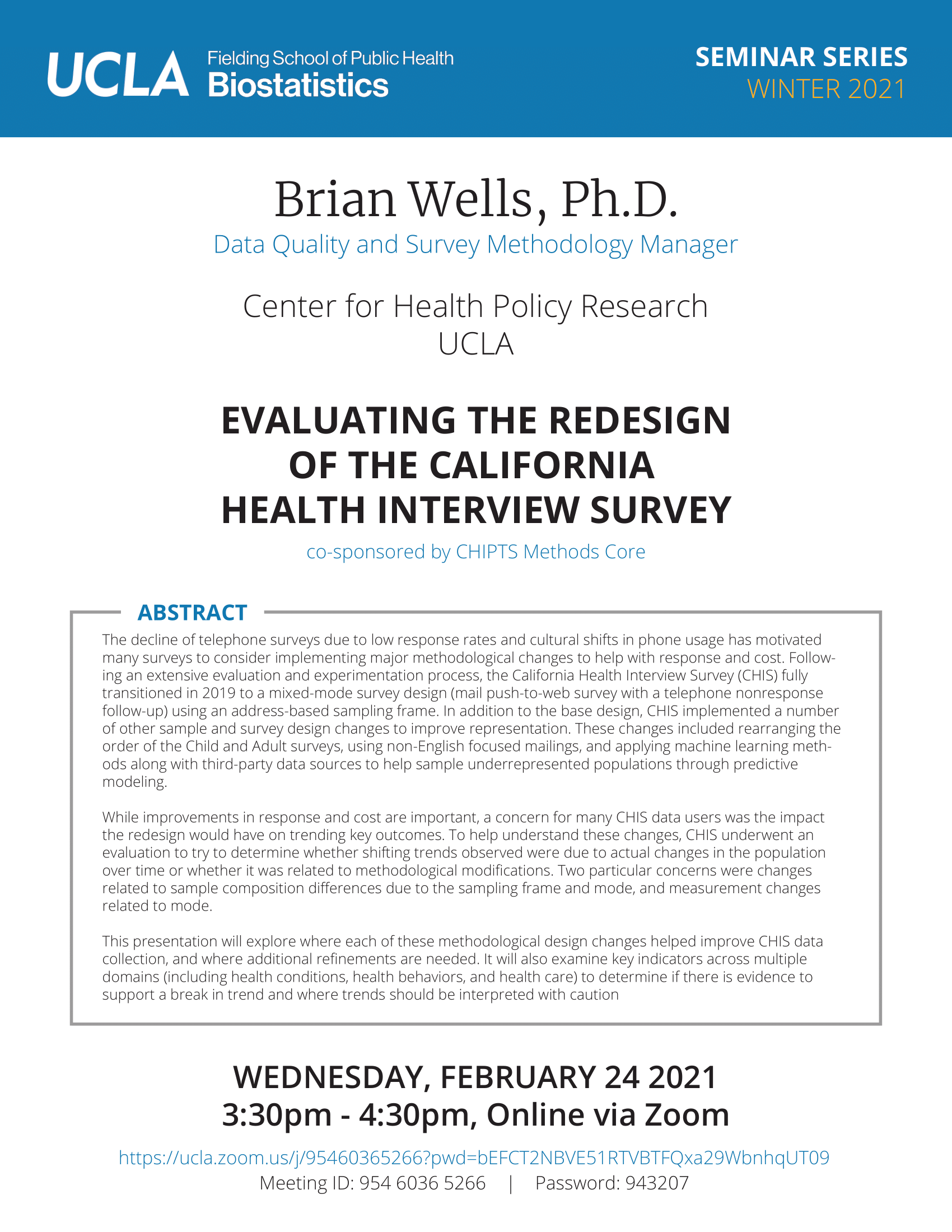
Brian Wells - 2/24/21
-
Jin Zhou - 2/22/21
Genetics of Within-Subject Variability and Diabetes Complications | UCLA CTSI Biostatistics Seminar
Date
Monday, February 22, 2021Time
12:00pm - 1:00pmLocation
Online via Zoom
https://uclahs.zoom.us/j/780633065Speaker
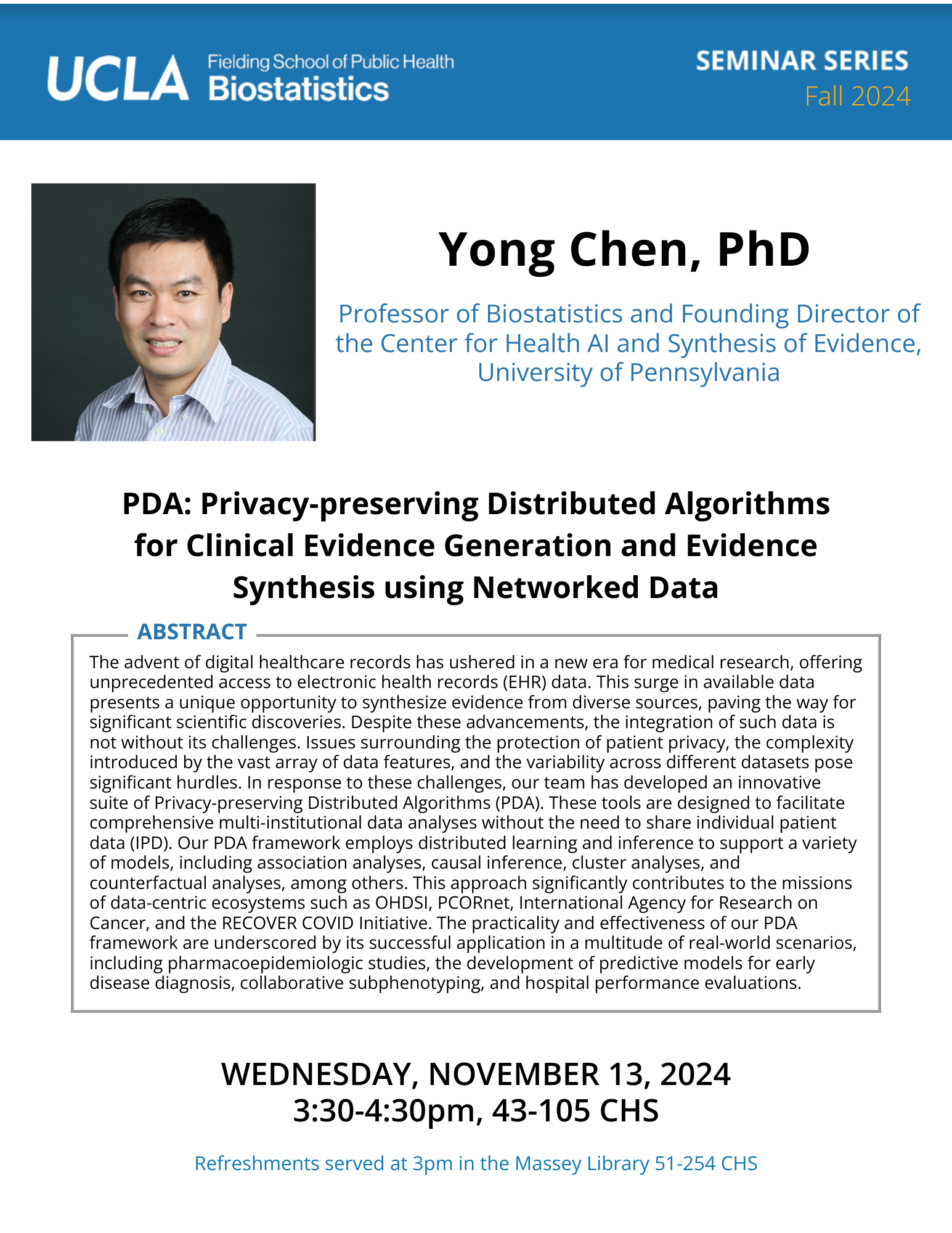
Jin Zhou, Ph.D.
Associate Professor, University of ArizonaAbstract
The development of diabetes complications, both macrovascular and microvascular, is heterogeneous, even when patients have the same glucose control and clinical features. Research searching for susceptible genes underlying diabetes complications is limited due to the complexities of studying diseases (complications) within a disease (diabetes). Our prior findings highlighted the importance of time-varying within subject (WS) glycemic (GV) and blood pressure variability (BPv) for developing diabetes complications. We hypothesize that genetic variants contribute to within-subject GV and BPv, contributing to progression to diabetes complications. In this talk, to quantify the genetic contributions to GV and BPv using biobank scale data, we develop a WS variance estimator by robust regression to estimate and inference the effects of both time-varying and time-invariant predictors on WS variance. Our method is robust against the distributional misspecification. We further boost the computational efficiency by implementing a score test that only needs to fit the null model once for the entire data sets, making it applicable to massive biobank data. We apply our method (vGWAS) to longitudinal glycemic, and blood pressure (BP) measures extracted from electronic medical records from UK Biobank. Our results complement current BP GWAS and shed light on disease mechanisms. -
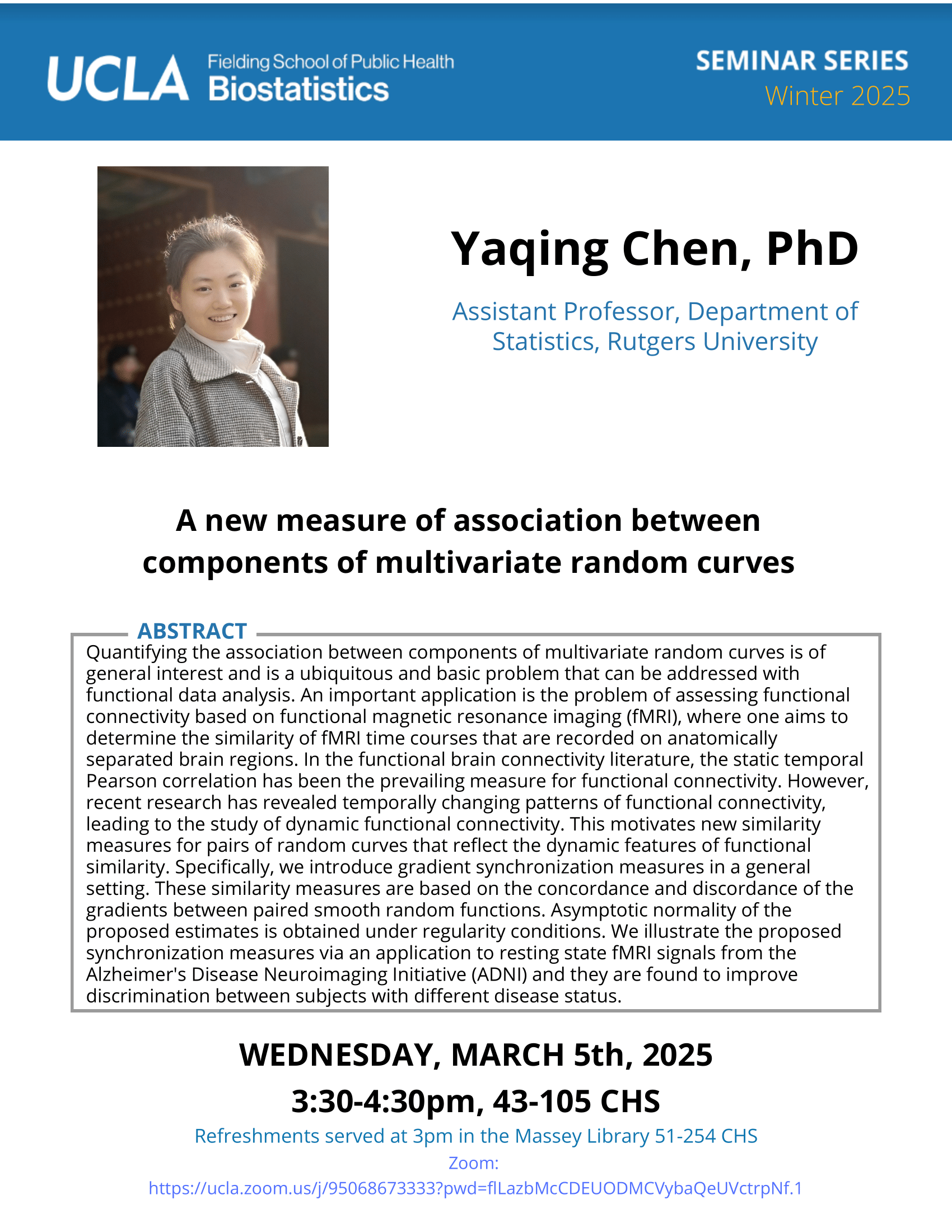
Vladimir Minin - 2/17/21
-
Abdelmonem Afifi - 2/10/21

-
Michele Peruzzi - 2/3/21
-
Guido Montufar - 1/20/21
-
Alexander Petersen - 1/13/21
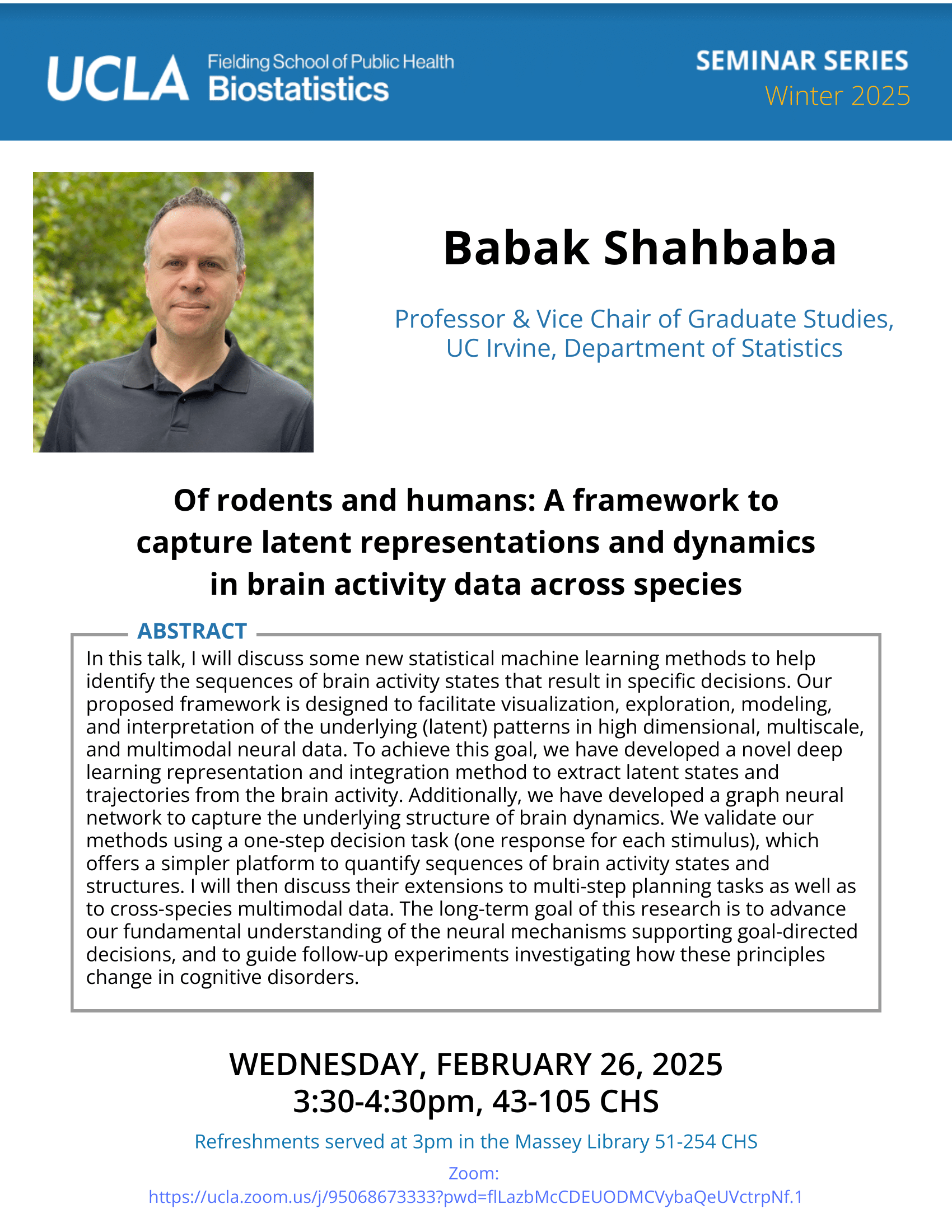
Fall 2020
-
Jelena Bradic - 12/9/20
Biostat Seminar: Causal Learning: excursions in double robustness
Date
Wednesday, December 9, 2020Time
3:30pm - 4:30pmLocation
Online via Zoom
https://ucla.zoom.us/j/97619833513?pwd=dVdpYUtIOFBaWk5TR0xkNktTVCt3UT09
Meeting ID: 976 1983 3513 | Passcode: 828359Speaker
Jelena Bradic
Associate Professor
Department of Mathematics & Halicioglu Data Science Institute
UC San DiegoAbstract
Recent progress in machine learning provides many potentially effective tools to learn estimates or make predictions from datasets of ever-increasing sizes. Can we trust such tools in clinical and highly-sensitive systems? If a learning algorithm predicts an effect of a new policy to be positive, what guarantees do we have concerning the accuracy of this prediction? The talk introduces new statistical ideas to ensure that the learned estimates satisfy some fundamental properties: especially causality and robustness. The talk will discuss potential connections and departures between causality and robustness. -
Noah Simon - 12/2/20
Biostat Seminar: Reframing proportional-hazards modeling for large time-to-event datasets with applications to deep learning
Date
Wednesday, December 2, 2020Time
3:30pm - 4:30pmLocation
Online via Zoom
https://ucla.zoom.us/j/98576333860?pwd=QTdSdmVZOWMwaHZscldJZG1GUzhBQT09
Meeting ID: 985 7633 3860 | Passcode: 140409Speaker
Noah Simon
Associate Professor
Department of Biostatistics
University of WashingtonAbstract
To build inferential or predictive survival models, it is common to assume proportionality of hazards and fit a model by maximizing the partial likelihood. This has been combined with non-parametric and high dimensional techniques, eg. spline expansions and penalties, to flexibly build survival models. New challenges require extension and modification of that approach. In a number of modern applications there is interest in using complex features such as images to predict survival. In these cases, it is necessary to connect more modern backends to the partial likelihood (such as deep learning infrastructures based on eg. convolutional/recurrent neural networks). In such scenarios, large numbers of observations are needed to train the model. However, in cases where those observations are available, the structure of the partial likelihood makes optimization difficult (if not completely intractable).In this talk we show how the partial likelihood can be simply modified to easily deal with large amounts of data. In particular, with this modification, stochastic gradient- based methods, commonly applied in deep learning, are simple to employ. This simplicity holds even in the presence of left truncation/right censoring. This can also be applied relatively simply with data stored in a distributed manner.
-
Annie Qu - 11/25/20
Biostat Seminar: Individualized Multi-directional Variable Selection
Date
Wednesday, November 25, 2020Time
3:30pm - 4:30pmLocation
Online via Zoom
https://ucla.zoom.us/j/99467473438?pwd=UmRadlpuR1pFaFpJY1hXN0h5WjNTQT09
Meeting ID: 994 6747 3438 | Passcode: 254642Speaker
Annie Qu
Professor
Department of Statistics
University of California, IrvineAbstract
In this talk we propose a heterogeneous modeling framework which achieves individual-wise feature selection and individualized covariates’ effects subgrouping simultaneously. In contrast to conventional model selection approaches, the new approach constructs a separation penalty with multi-directional shrinkages, which facilitates individualized modeling to distinguish strong signals from noisy ones nd selects different relevant variables for different individuals. Meanwhile, the proposed model identifies subgroups among which individuals share similar covariates’ effects, and thus improves individualized estimation efficiency and feature selection accuracy. Moreover, the proposed model also incorporates within-individual correlation for longitudinal data to gain extra efficiency. We provide a general theoretical foundation under a double-divergence modeling framework where the number of individuals and the number of individual-wise measurements can both diverge, which enables inference on both an individual level and a population level. In particular, we establish strong oracle property for the individualized estimator to ensure its optimal large sample property under various conditions. -
Lu Tian - 11/18/20
Biostat Seminar: Constructing Confidence Interval for RMST under Group Sequential Setting
Date
Wednesday, November 18, 2020Time
3:30pm - 4:30pmLocation
Online via Zoom
https://ucla.zoom.us/j/94791678395?pwd=aTZoV0trQzdIaEE5WjNLY0IzSElpZz09
Meeting ID: 947 9167 8395 | Passcode: 042094Speaker
Lu Tian
Associate Professor of Biomedical Data Science and Statistics
Stanford UniversityAbstract
It is appealing to compared survival distributions based on restricted mean survival time (RMST), since it generates a clinically interpretable summary of the treatment effect and can be estimated nonparametrically without assuming restrictive model assumptions such as the proportional hazards assumption. However, there are special challenges in designing and analyzing group sequential study based on RMST, because the truncation timepoint of the RMST in the interim analysis often differs from that in the final analysis. A valid test controls the unconditional type one error has been developed in the past. However, there is no appropriate statistical procedure for constructing the confidence interval for the treatment effect measured by a contrast in RMST, while it is crucial for informative clinical decision making. In this talk, I will review some important design issues for study based on RMST. I will then discuss how to conduct hypothesis testing and how to construct confidence intervals for the difference RMST in a group sequential setting. Examples and numerical studies will be presented to illustrate the method. -
Biostatistics Admission Committee - 11/10/20
Biostatistics Admission Information Session
Date
Tuesday, November 10, 2020Time
1:00pm - 2:00pmLocation
Online via Zoom
https://ucla.zoom.us/j/94445671105?pwd=emxESzJiUXNxTHJjTkRGK0JSVm8yZz09
Meeting ID: 944 4567 1105 | Passcode: 742314Details
Biostatistics admission committee members and biostatistics student representatives are available to answer any questions you have regarding graduate programs (MS, MPH, PhD) in Biostatistics. -
Oscar Madrid Padilla - 11/4/20
Biostat Seminar: Optimal post-selection inference for sparse signals: a nonparametric empirical-Bayes
Date
Wednesday, November 4, 2020Time
3:30pm - 4:30pmLocation
Online via Zoom
https://ucla.zoom.us/j/93327847797?pwd=OVpGUTg1SEJGZ1VWb3ZJK08rRThrZz09
Meeting ID: 933 2784 7797 | Passcode: 001430Speaker
Oscar Hernan Madrid Padilla
Assistant Professor
Statistics Department, UCLAAbstract
Many recently developed Bayesian methods have focused on sparse signal detection. However, much less work has been done addressing the natural follow-up question: how to make valid inferences for the magnitude of those signals after selection. Ordinary Bayesian credible intervals suffer from selection bias, owing to the fact that the target of inference is chosen adaptively. Existing Bayesian approaches for correcting this bias produce credible intervals with poor frequentist properties, while existing frequentist approaches require sacrificing the benefits of shrinkage typical in Bayesian methods, resulting in confidence intervals that are needlessly wide. We address this gap by proposing a nonparametric empirical-Bayes approach for constructing optimal selection-adjusted confidence sets. Our method produces confidence sets that are as short as possible on average, while both adjusting for selection and maintaining exact frequentist coverage uniformly over the parameter space. Our main theoretical result establishes an important consistency property of our procedure: that under mild conditions, it asymptotically converges to the results of an oracle-Bayes analysis in which the prior distribution of signal sizes is known exactly. Across a series of examples, the method outperforms existing frequentist techniques for post selection inference, producing confidence sets that are notably shorter but with the same coverage guarantee.
Winter 2020
-
Murali Haran - 3/4/20 (CANCELLED)
(CANCELLED) | Biostat Seminar: Inference in the Presence of Intractable Normalizing Functions
Date
Wednesday, March 4, 2020Time
3:30pm - 4:30pmRefreshments at 3:00pm in 51-254 CHS
Location
33-105 CHSSpeaker
Murali Haran
Professor and Head, Department of Statistics
Penn State UniversityAbstract
Models with intractable normalizing functions arise frequently in statistics. Common examples of such models include exponential random graph models for social networks and Markov point processes for ecology and disease modeling. Inference for these models is complicated because the normalizing functions of their probability distributions include the parameters of interest. We provide a framework for understanding existing algorithms for Bayesian inference for these models, comparing their computational and statistical efficiency, and discussing their theoretical bases. We propose an algorithm that provides computational gains over existing methods by replacing Monte Carlo approximations to the normalizing function with a Gaussian process-based approximation. We provide theoretical justification for this method. We also develop a closely related algorithm that is applicable more broadly to any likelihood function that is expensive to evaluate. We illustrate the application of our methods to a variety of challenging simulated and real data examples, including an exponential random graph model, a Markov point process, and a model for infectious disease dynamics. Our algorithms show significant gains in computational efficiency over existing methods, and have the potential for greater gains for more challenging problems. For a random graph model example, this gain in efficiency allows us to carry out Bayesian inference when other algorithms are computationally impractical. -
Tse L. Lai - 2/26/20
Biostat Seminar: Real-world Evidence in Drug Development and Regulatory Submission
Date
Wednesday, February 26, 2020Time
3:30pm - 4:30pmRefreshments at 3:00pm in 51-254 CHS
Location
33-105A CHSSpeaker
Tse L. Lai
Ray Lyman Wilbur Professor of Statistics
Stanford UniversityAbstract
There has been growing interest in using real-world data (RWD) and evidence (RWE) for drug development since the passage of the 21st Century Cures Act in Dec 2016. The US FDA released its Framework for Real-World Evidence Program in Dec 2018 and subsequently issued a draft guidance for industry on submitting documents using RWD & E for drugs and biologics. I will discuss statistical challenges and opportunities in using RWD/RWE for drug development and regulatory submission, and describe some ongoing projects that are summarized in my forthcoming book (Chapman & Hall/CRC, 2020) with Richard Baumgartner and Jie Chen of Merck on RWD & E. -
Dennis K.J. Lin - 2/19/20
Biostat Seminar: Ghost Data
Date
Wednesday, February 19, 2020Time
3:30pm - 4:30pmRefreshments at 3:00pm in 51-254 CHS
Location
33-105 CHSSpeaker
Dennis K.J. Lin
University Distinguished Professor
Department of Statistics, The Pennsylvania State University, University Park, USAAbstract
As natural as the real data, ghost data is everywhere—it is just data that you cannot see. We need to learn how to handle it, how to model with it, and how to put it to work. Some examples of ghost data are (see, Sall, 2017):
(a) Virtual data—it isn’t there until you look at it;
(b) Missing data—there is a slot to hold a value, but the slot is empty;
(c) Pretend data—data that is made up;
(d) Highly Sparse Data—whose absence implies a near zero, and
(e) Simulation data—data to answer “what if.”
For example, absence of evidence/data is not evidence of absence. In fact, it can be evidence of something. More Ghost Data can be extended to other existing areas: Hidden Markov Chain, Two-stage Least Square Estimate, Optimization via Simulation, Partition Model, Topological Data, just to name a few. Three movies will be discussed in this talk: (1) “The Sixth Sense” (Bruce Wallis)—I can see things that you cannot see; (2) “Sherlock Holmes” (Robert Downey)—absence of expected facts; and (3) “Edge of Tomorrow” (Tom Cruise)—how to speed up your learning (AlphaGo-Zero will also be discussed). It will be helpful, if you watch these movies before coming to my talk. This is an early stage of my research in this area–any feedback from you is deeply appreciated. Much of the basic idea is highly influenced via Mr. John Sall (JMP-SAS). -
Sean Jewell - 2/5/20
Biostat Seminar: Estimation and Inference for Changepoint Models
Date
Wednesday, February 5, 2020Time
3:30pm - 4:30pmRefreshments at 3:00pm in 51-254 CHS
Location
33-105A CHSSpeaker
Sean Jewell
PhD Candidate
Department of Statistics at the University of WashingtonAbstract
This talk is motivated by statistical challenges that arise in the analysis of calcium imaging data, a new technology in neuroscience that makes it possible to record from huge numbers of neurons at single-neuron resolution. In the first part of this talk, I will consider the problem of estimating a neuron’s spike times from calcium imaging data. A simple and natural model suggests a non-convex optimization problem for this task. I will show that by recasting the non-convex problem as a changepoint detection problem, we can efficiently solve it for the global optimum using a clever dynamic programming strategy.In the second part of this talk, I will consider quantifying the uncertainty in the estimated spike times. This is a surprisingly difficult task, since the spike times were estimated on the same data that we wish to use for inference. To simplify the discussion, I will focus specifically on the change-in-mean problem, and will consider the null hypothesis that there is no change in mean associated with an estimated changepoint. My proposed approach for this task can be efficiently instantiated for changepoints estimated using binary segmentation and its variants, L0 segmentation, or the fused lasso. Moreover, this framework allows us to condition on much less information than existing approaches, thereby yielding higher-powered tests. These ideas can be easily generalized to the spike estimation problem.
This talk will feature joint work with Toby Hocking, Paul Fearnhead, and Daniela Witten.
-
Jessica Gronsbell - 2/3/20
Biostat Seminar: Estimation and Inference for Changepoint Models
Date
Monday, February 3, 2020Time
3:30pm - 4:30pmRefreshments at 3:00pm in 51-254 CHS
Location
63-105A CHSSpeaker
Jessica Gronsbell, PhD
Data Scientist
Alphabet’s Verily Life SciencesAbstract
The widespread adoption of electronic health records (EHR) and their subsequent linkage to specimen biorepositories has generated massive amounts of routinely collected medical data for use in translational research. These integrated data sets enable real-world predictive modeling of disease risk and progression. However, data heterogeneity and quality issues impose unique analytical challenges to the development of EHR-based prediction models. For example, ascertainment of validated outcome information, such as presence of a disease condition or treatment response, is particularly challenging as it requires manual chart review. Outcome information is therefore only available for a small number of patients in the cohort of interest, unlike the standard setting where this information is available for all patients. In this talk I will discuss semi-supervised and weakly-supervised learning methods for predictive modeling in such constrained settings where the proportion of labeled data is very small. I demonstrate that leveraging unlabeled examples can improve the efficiency of model estimation and evaluation and in turn substantially reduce the amount of labeled data required for developing prediction models. -
Wesley Tansey - 1/31/20
Biostat Seminar: Modeling and testing in high-throughput cancer drug screenings
Date
Friday, January 31, 2020Time
11:00am - 12:00pmRefreshments at 10:30am in 51-254 CHS
Location
33-105A CHSSpeaker
Wesley Tansey
Postdoctoral Research Scientist
Columbia UniversityAbstract
High-throughput drug screens enable biologists to test hundreds of candidate drugs against thousands of cancer cell lines. The sensitivity of a cell line to a drug is driven by the molecular features of the tumor (e.g. gene mutations and expression). In this talk, I will consider two scientific goals at the forefront of cancer biology: (i) predicting drug response from molecular features, and (ii) discovering gene-drug associations that represent candidates for future drug development. I will present an end-to-end model of cancer drug response that combines hierarchical Bayesian modeling with deep neural networks to learn a flexible function from molecular features to drug response. The model achieves the first goal of state-of-the-art predictive performance, but the black box nature of deep learning makes the model difficult to interpret, presenting a barrier to the second goal of uncovering gene-drug associations. I will use this challenge as motivation for the development of a new method, the holdout randomization test (HRT), for conditional independence testing with black box predictive models. Applying the HRT to the deep probabilistic model of cancer drug response yields more biologically-plausible gene-drug associations than the current analysis technique in biology. I will use these projects to illustrate how statisticians can work closely with biologists to create a virtuous cycle where cutting -edge experiments lead to new statistical models and methods, which in turn drive all of science forward. -
Zhengwu Zhang - 1/27/20
Biostat Seminar: Statistical Analysis of Brain Structural Connectomes
Date
Monday, January 27, 2020Time
3:30pm - 4:30pmRefreshments at 3:00pm in 51-254 CHS
Location
63-105A CHSSpeaker
Zhengwu Zhang
Assistant Professor
University of RochesterAbstract
There have been remarkable advances in imaging technology, used routinely and pervasively in many human studies, that non-invasively measures human brain structure and function. Among them, a particular imaging modality called diffusion magnetic resonance imaging (dMRI) is used to infer shapes of millions of white matter fiber tracts that act as highways for neural activity and communication across the brain. The collection of interconnected fiber tracts is referred to as the brain connectome. There is increasing evidence that an individual’s brain connectome plays a fundamental role in cognitive functioning, behavior, and the risk of developing mental disorders. Improved mechanistic understanding of relationships between brain connectome structure and phenotypes is critical to the prevention and treatment of mental disorders. However, progress in this area has been limited duo to the complexity of the data. In this talk, I will present challenges of analyzing such data and our recent progress, including connectome reconstruction and novel statistical modeling methods. -
Andrew Holbrook - 1/22/20
Biostat Seminar: Bayes in the time of Big Data
Date
Wednesday, January 22, 2020Time
3:30pm - 4:30pmRefreshments at 3:00pm in 51-254 CHS
Location
33-105 CHSSpeaker
Andrew Holbrook
Postdoctoral Scholar
UCLA, Human GeneticsAbstract
Big Bayes is the computationally intensive co-application of big data and large, expressive Bayesian models for the analysis of complex phenomena in scientific inference and statistical learning. Standing as an example, Bayesian multidimensional scaling (MDS) can help scientists learn viral trajectories through space and time, but its computational burden prevents its wider use. Crucial MDS model calculations scale quadratically in the number of observations. We mitigate this limitation through massive parallelization using multi-core central processing units, instruction-level vectorization and graphics processing units (GPUs). Fitting the MDS model using Hamiltonian Monte Carlo, GPUs can deliver more than 100-fold speedups over serial calculations and thus extend Bayesian MDS to a big data setting. To illustrate, we employ Bayesian MDS to infer the rate at which different seasonal influenza virus subtypes use worldwide air traffic to spread around the globe. We examine 5392 viral sequences and their associated 14 million pairwise distances arising from the number of commercial airline seats per year between viral sampling locations. To adjust for shared evolutionary history of the viruses, we implement a phylogenetic extension to the MDS model and learn that subtype H3N2 spreads most effectively, consistent with its epidemic success relative to other seasonal influenza subtypes. -
Lan Luo - 1/15/20
Biostat Seminar: Bayes in the time of Big Data
Date
Wednesday, January 15, 2020Time
3:30pm - 4:30pmRefreshments at 3:00pm in 51-254 CHS
Location
33-105 CHSSpeaker
Lan Luo
Doctoral Candidate
Department of Biostatistics
University of Michigan, Ann ArborAbstract
This research is largely motivated by the challenges in modeling and analyzing streaming health data, which are becoming increasingly popular data sources in the fields of biomedical science and public health. In this work, the term “streaming data” refers to high throughput recording of large volumes of observations collected sequentially and perpetually over time, such as national disease registry, mobile health, and disease surveillance. Due to the large volume and frequent updates intrinsic to this type of data, major challenges arising from the analysis of streaming data pertain to data storage and information updating. This talk primarily concerns the development of a real-time statistical estimation and inference method for regression analysis, with a particular objective of addressing challenges in streaming data storage and computational efficiency. Termed as “renewable estimation”, this method greatly helps overcome the data sharing barrier, reduce data storage cost, and improve computing speed, all without loss of statistical efficiency. The proposed algorithms for streaming real-time regression will be demonstrated in generalized linear models (GLM) for cross-sectional data. I will discuss both conceptual understanding and theoretical guarantees of the renewable method and illustrate its performance via numerical examples. This is joint work with my supervisor Peter Song at the University of Michigan. -
Allison Meisner - 1/13/20
Biostat Seminar: Risk Models with Polygenic Risk Scores
Date
Monday, January 13, 2020Time
3:30pm - 4:30pmRefreshments at 3:00pm in 51-254 CHS
Location
63-105 CHSSpeaker
Allison Meisner, PhD
Postdoctoral Fellow
Department of Biostatistics
Johns Hopkins UniversityAbstract
Most complex diseases are the result of environmental variables, genetic factors, and their interaction. In building risk models, it is important to account for each of these components to enable estimation of risk and identification of high-risk subgroups. Historically, research into the genetic determinants of disease has largely focused on the role of individual variants. However, this endeavor is complicated by the fact that most diseases are highly polygenic and result from the combined effect of many variants, each with small effect. A great deal of attention has been paid recently to polygenic risk scores, which represents the total genetic burden of a given trait. Here, I present recent work on utilizing polygenic risk scores in risk models, alongside environmental risk factors. This includes an efficient case-only method for using polygenic risk scores to identify gene- environment interactions and an expansive analysis of the combined utility of polygenic risk scores for specific diseases and mortality risk factors in predicting survival in the UK Biobank, a large cohort study. I will also touch on possibilities for future work in this area, including the use of polygenic risk scores in treatment selection.