UCLA Fielding faculty drive improved methods to research spread of infectious viral diseases
Peer-reviewed studies detail use of techniques to better track the spread of disease and highlight UCLA Fielding’s work on public health methodology.
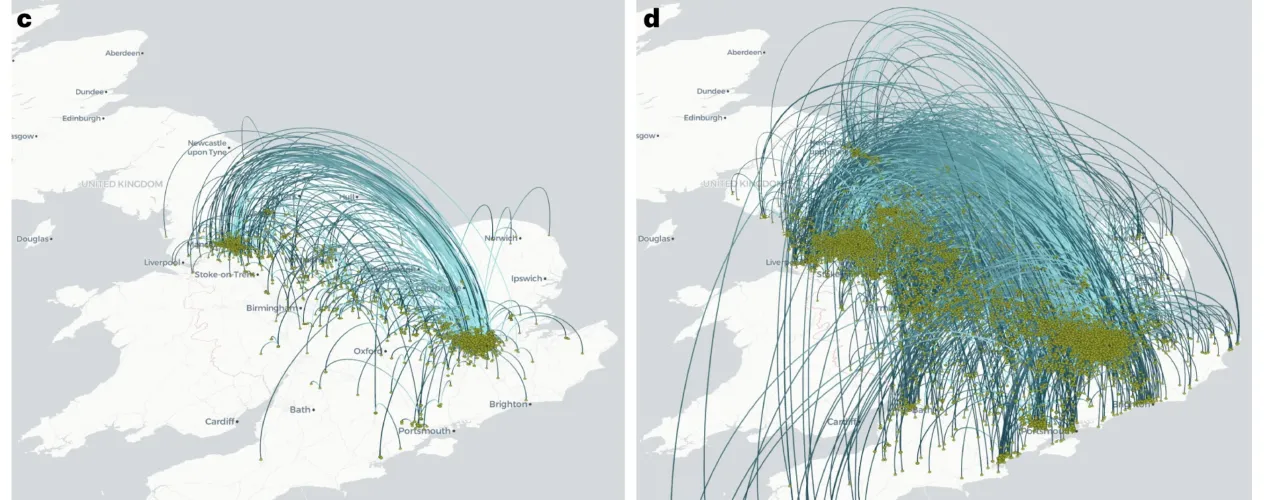
Two related and recently published studies by international teams - including researchers with the UCLA Fielding School of Public Health - have identified methods to improve how infectious diseases, including COVID-19, mpox, and Ebola, can be tracked and better understood by public health officials in an emergency.
“The results enable us to write clear guidelines for the use of novel computational approaches analyzing viral sequences to guide public health decisions in emerging infectious disease crises,” said Dr. Marc Suchard, a physician and professor in the UCLA Fielding School of Public Health's Department of Biostatistics. “Our open-source methods are available to the scientific community and can be used to investigate the drivers of viral spread through space and between people to design tailored intervention strategies.”
The studies focus on the use of viral disease genomes – the genetic material contained within a virus particle, which can be composed of either deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) - to estimate the dispersal history of the virus responsible for an epidemic, a task known as phylogeographic inference. The researchers developed and evaluated the performance of three new analytical approaches using standardized software to create phylogeographic reconstructions, improving understanding of how quickly a virus can disperse across a given population.
As an example of how the techniques could be used, the researchers used historical data from the 2021-22 COVID-19 (SARS-CoV-2) outbreak in the United Kingdom and used the newly developed methods to check if the dispersal patterns could have been discerned earlier than they were, historically. Overall, the new techniques improved the speed of getting results of the analyses, in some cases by up to 300-400 times, researchers said.
“The usefulness of this sort of expedited analysis in improving a public health department’s reaction to an emerging public health crisis is obvious,” said UCLA Fielding’s Dr. Andrew Holbrook, an assistant professor in the Department of Biostatistics and a study co-author. “The more, and the earlier, those dealing with an outbreak understand what is happening, the more likely it can be slowed or even stopped.”
The two peer-reviewed studies, all by researchers affiliated with UCLA and universities or research institutes in the United States, Belgium, France, the Netherlands, New Zealand, and the United Kingdom, are:
- “BEAST X for Bayesian phylogenetic, phylogeographic and phylodynamic inference,” in the London-based journal Nature Methods, in July, 2025; and
- “Comparative performance of viral landscape phylogeography approaches,” in the Washington, DC-based journal the Proceedings of the National Academy of Sciences (PNAS), in June, 2025.
“Recent innovations in statistics are quickly changing the scale of infectious disease research, and I am excited that they are both shaping my education at UCLA and impacting the world around us,” said study co-author Yucai Shao, a doctoral student at UCLA Fielding.
This work, conducted by integrated teams of researchers from the United States and internationally, demonstrate the real-world impact of research led by UCLA scholars, and how advances in data science can be used to improve tools used by public health officials, experts said.
“The impact of innovative research led by UCLA Fielding faculty and students extends far beyond university walls,” said Dr. Ron Brookmeyer, dean of the UCLA Fielding School of Public Health and a distinguished professor of biostatistics. “Transformative findings and solutions reach individuals and communities and contribute to building a healthier future for all.”
Funding
The work published in Nature received support from the U.S. National Science Foundation (NSF) and the U.S. National Institutes of Health (NIH), as well as the European Union, the Research Foundation – Flanders, the Bill & Melinda Gates Foundation, and from the universities and institutions where the authors are affiliated. The work published in PNAS further received support from the European Union, foundations and governments in Belgium, France, and the Netherlands, as well as the universities and institutions where the authors are affiliated.
The funders, including the U.S. NSF and U.S. NIH, had no role in the conduct of the studies or in the decision to publish findings. The U.S. government grants were all awarded competitively.

