Research helps identify high-risk populations to aid health officials combating the pandemic
UCLA researchers have developed a method to better guide public policy related to the control and prevention of COVID-19.
A team of UCLA Fielding School of Public Health researchers has developed analyses to identify those most at risk from the pandemic, using demographic data to estimate test-based infection and test-based case fatality rates across California.
The research, published in the December edition of the peer-reviewed journal Epidemics, introduces a method that combines aggregated publicly available COVID-19 case and fatality data with population demographic survey data. This helps to overcome the limitations of aggregated summary statistics and yields estimates of COVID-19 test-based infection and case fatality rates across different subgroup population demographics— key quantities for guiding public policy related to the control and prevention of COVID-19.
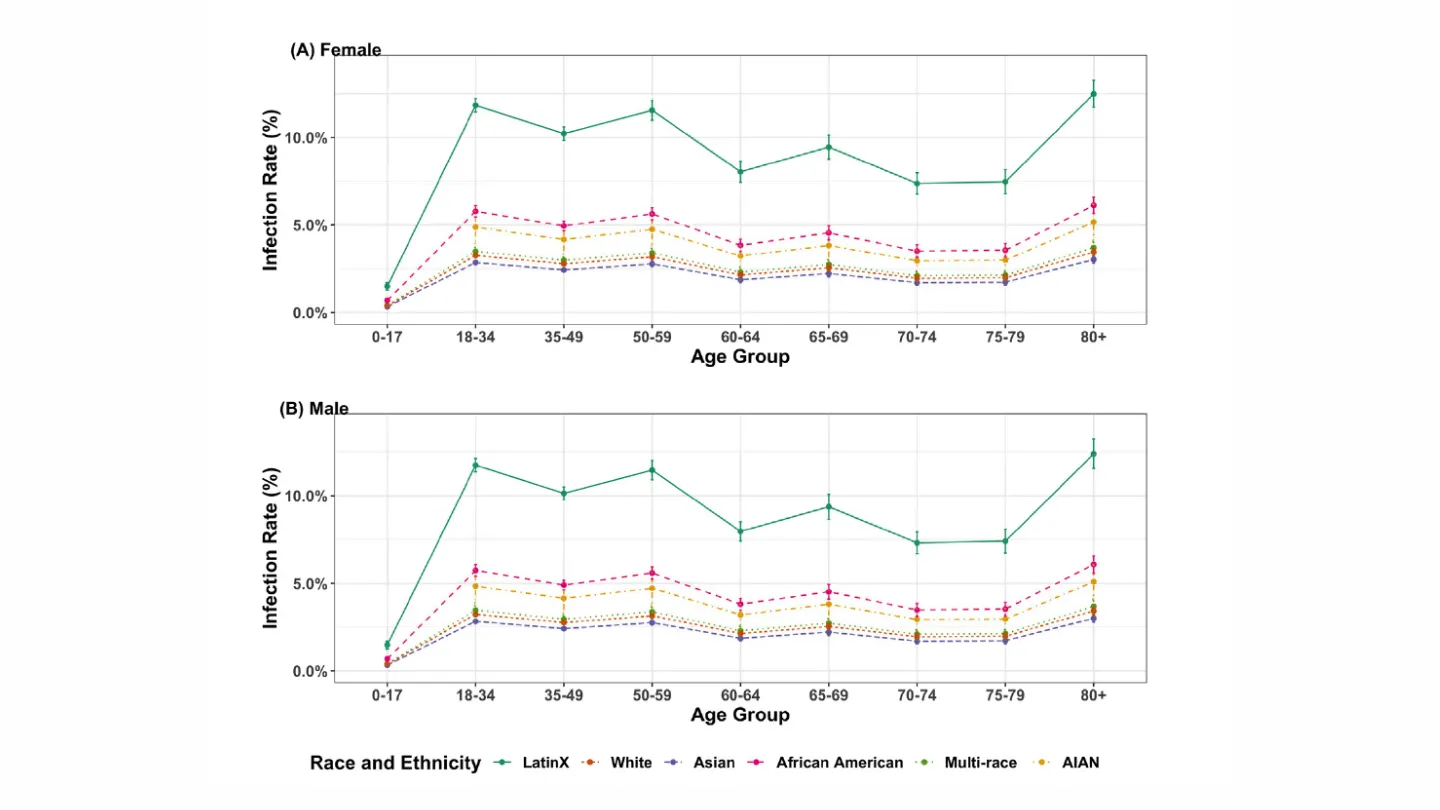
Fig. 2. Estimated test-based infection rates given age and race/ethnicity, stratified by gender. ((A) and (B) present the bootstrapped mean infection rates for female and male respectively. Only 6 racial and ethnicity groups are considered in the figures, including LatinX/ Hispanic (LatinX), White/ Caucasian (White), Asian, African American/Black (AA), Multi-Race, and American Indian or Alaska Native (AIAN). The overall infection rate was assumed to be 5.2%, and the error bars denote two bootstrap standard errors.).
“In emerging epidemics, early estimates of key epidemiological characteristics of the disease are critical for guiding public policy, and identifying high-risk population subgroups aids policymakers and health officials in combating the epidemic,” said Christina Ramirez, UCLA Fielding School of Public Health professor of biostatistics. “This has been challenging during the pandemic because governmental agencies typically release aggregate COVID-19 data as summaries; these may identify broad disparities in outcomes, but typically do not provide granular data that would include combinations of demographic characteristics such as age, race and gender.”
The researchers used daily COVID-19 data for California from the California Department of Public Health (CDPH) and the 2017–2018 wave of the California Health Interview Survey (CHIS), a statewide research project by the Fielding School’s UCLA Center for Health Policy Research (CHPR).
“Our approach combines aggregate COVID-19 case and fatality data with population-level demographic survey data to estimate test-based infection and case fatality rates for population subgroups across combinations of demographic characteristics,” said co-author Dr. Marc Suchard, UCLA Fielding School of Public Health professor of biostatistics. “What is shows is that as tragic as the pandemic has been for Californians generally, it has hit certain groups even harder.”
The team’s analysis indicates that in California, males have higher test-based infection rates and test-based case fatality rates across age and race/ethnicity groups, with the gender gap widening with increasing age. Although elderly infected with COVID-19 are at an elevated risk of mortality, the test-based infection rates do not always increase with age in a linear fashion.
“The workforce population with ages from 18–59 have a higher infection rate comparing with children, adolescents, and other senior citizens, except for people in their 80 and above,” Ramirez said. “We also found that the elevated infection and mortality risk for males and greater mortality risk for all races increase with age.”
The subgroups with the highest five test-based case fatality rates are all-male groups with race as African American, Asian, Multi-race, Latino, and White, followed by African American females, indicating that African Americans are an especially vulnerable California subpopulation.
Sponsorship and Funding:
Support was provided by the authors’ institutions.
Data Availability Statement:
All relevant data are publicly available from the California Department of Public Health and the California Health Interview Survey.
Citation:
Di Xiong, Lu Zhang, Gregory L. Watson, Phillip Sundin, Teresa Bufford, Joseph A. Zoller, John Shamshoian, Marc A. Suchard, Christina M. Ramirez, “Pseudo-likelihood based logistic regression for estimating COVID-19 infection and case fatality rates by gender, race, and age in California,” in Epidemics, Volume 33, 2020, 100418, ISSN 1755-4365,
https://doi.org/10.1016/j.epidem.2020.100418.
The UCLA Fielding School of Public Health, founded in 1961, is dedicated to enhancing the public's health by conducting innovative research, training future leaders and health professionals from diverse backgrounds, translating research into policy and practice, and serving our local communities and the communities of the nation and the world. The school has 631 students from 26 nations engaged in carrying out the vision of building healthy futures in greater Los Angeles, California, the nation and the world.
Attachment: Epidemics Ramirez et alFaculty Referenced by this Article

Automated and accessible artificial intelligence methods and software for biomedical data science.